Study Identifies Potential Gene Targets to Overcome Drug Resistance in Breast Cancer
Researchers developed a computational framework to pinpoint metabolic gene targets that can re-sensitize drug-resistant breast cancer cells. By simulating human metabolism, the study identified specific genes that, when suppressed, could make resistant cancer cells responsive to treatment again. This approach not only holds promise for cancer therapies but also for treating metabolic diseases like diabetes.
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
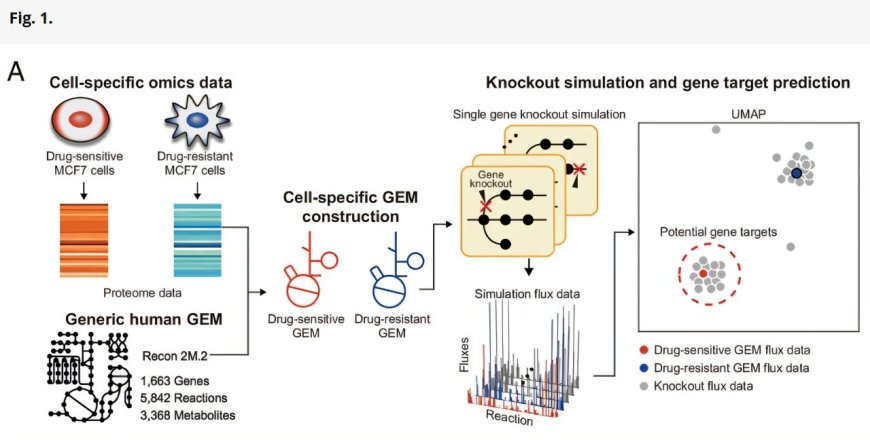
Computational framework that can identify metabolic gene targets to revert the metabolic state of the drug-resistant cells to that of the drug-sensitive parental cells. Credit: Proceedings of the National Academy of Sciences (2025). DOI: 10.1073/pnas.2425384122
One of the biggest obstacles in cancer treatment is drug resistance in cancer cells. Conventional efforts have focused on identifying new drug targets to eliminate these resistant cells, but such approaches can often lead to even stronger resistance.
A research team led by Professors Hyun Uk Kim and Yoosik Kim from the Department of Chemical and Biomolecular Engineering developed a computational framework that predicts metabolic gene targets to re-sensitize drug-resistant breast cancer cells. This was achieved using a metabolic network model capable of simulating human metabolism.
This technique holds promise not only for a variety of cancer therapies but also for treating metabolic diseases such as diabetes.
The study was published in Proceedings of the National Academy of Sciences.
Focusing on metabolic alterations—key characteristics in the formation of drug resistance—the researchers developed a metabolism-based approach to identify gene targets that could enhance drug responsiveness by regulating the metabolism of drug-resistant breast cancer cells.
The team first constructed cell-specific metabolic network models by integrating proteomic data obtained from two different types of drug-resistant MCF7 breast cancer cell lines: one resistant to doxorubicin and the other to paclitaxel. They then performed gene knockout simulations on all of the metabolic genes and analyzed the results.
As a result, they discovered that suppressing certain genes could make previously resistant cancer cells responsive to anticancer drugs again. Specifically, they identified GOT1 as a target in doxorubicin-resistant cells, GPI in paclitaxel-resistant cells, and SLC1A5 as a common target for both drugs.
The predictions were experimentally validated by suppressing proteins encoded by these genes, which led to the re-sensitization of the drug-resistant cancer cells.
Furthermore, consistent re-sensitization effects were also observed when the same proteins were inhibited in other types of breast cancer cells that had developed resistance to the same drugs.
Professor Yoosik Kim said, \"Cellular metabolism plays a crucial role in various intractable diseases, including infectious and degenerative conditions. This new technology, which predicts metabolic regulation switches, can serve as a foundational tool not only for treating drug-resistant breast cancer but also for a wide range of diseases that currently lack effective therapies.\"
Professor Hyun Uk Kim, who led the study, emphasized, \"The significance of this research lies in our ability to accurately predict key metabolic genes that can make resistant cancer cells responsive to treatment again—using only computer simulations and minimal experimental data. This framework can be widely applied to discover new therapeutic targets in various cancers and metabolic diseases.\"
More information: JinA Lim et al, Genome-scale knockout simulation and clustering analysis of drug-resistant breast cancer cells reveal drug sensitization targets, Proceedings of the National Academy of Sciences (2025). DOI: 10.1073/pnas.2425384122 Journal information: Proceedings of the National Academy of Sciences
What's Your Reaction?
 Like
0
Like
0
 Dislike
0
Dislike
0
 Love
0
Love
0
 Funny
0
Funny
0
 Angry
0
Angry
0
 Sad
0
Sad
0
 Wow
0
Wow
0












































































































































































